Define your experimental data in this section. More then one file can be uploaded, each file representing data measured at different magnetic field. If data were measured at multiple field strengths, the residue types and numbers will be compared and chemical shift differences fitted globally to calculate kinetic parameters.
The input-file should have the following format:
| 1st line | Value of the B0 field for the nuclei of interest in MHz (e.g. 60.12). |
| 2nd line | Value of the constant relaxation time for the CPMG experiment in seconds (e.g. 0.04). |
| 3rd line | Comment line. In the example file, we described the data format (e.g. #nu_cpmg(Hz) R2(1/s) Esd(R2)). |
| > 3rd line |
The first line of the block defines the residue type and number and optionally the intrinsic relaxation rate R20 from HEROINE experiment. The following lines contain the CPMG frequencies ( 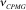 , s-1), relaxation rates ( , s-1), relaxation rates (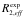 , s-1) and errors in relaxation rates ( , s-1) and errors in relaxation rates (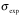 , s-1). , s-1).
|